RiboScanner
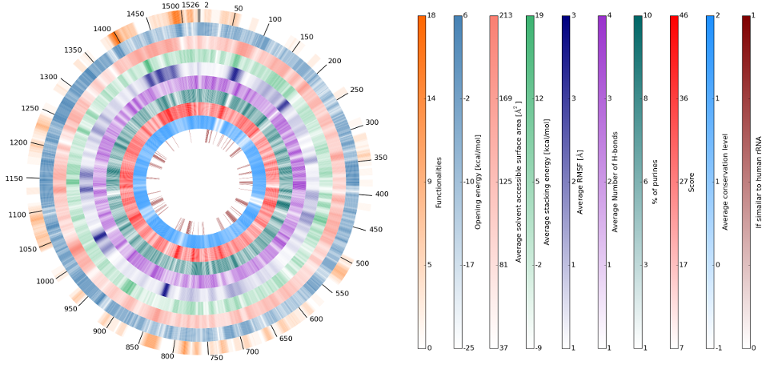
We have designed a server to aid in the search for putative binding sites in 16S ribosomal RNA that could be targeted by peptide nucleic acid oligomers. You can browse all of the computed characteristics via the interactive plots in 16S rRNA and 23S rRNA bookmarks. While looking for the best rRNA sites we have computed multiple different characteristics of rRNA. The entire procedure, analyses and results are described in our publication. Please cite A. Górska et al., Scanning of 16S RNA for peptide nucleic acid targets, J. Phys. Chem. B, doi:10.1021/acs.jpcb.6b2082 if you find the server useful.